Lesson 5. Convert R Markdown to PDF or HTML
In the previous tutorials we’ve learned about the R Markdown format and how
to create a report using R Markdown in RStudio. In this tutorial, we will
render or knit an R Markdown document to a web friendly, html format using
the R knitr package. knitr can be used to convert R Markdown files to many
different formats including: html, pdf, GitHub markdown (.md) and more.
Learning Objectives
At the end of this lesson, you will:
- Be able to produce (
knit) anhtmlfile from anR Markdownfile. - Know how to modify chuck options to change what is rendered and not rendered on the output
htmlfile.
What You Need
You will need the most current version of R and, preferably, RStudio loaded on
your computer to complete this tutorial. You will also need an R Markdown
document that contains a YAML header, code chunks and markdown segments.
Install R Packages
- knitr:
install.packages("knitr") - rmarkdown:
install.packages("rmarkdown")
What is Knitr?
knitr is the R package that we use to convert an R Markdown document into another,
more user friendly format like .html or .pdf.
The knitr package allows us to:
- Publish & share preliminary results with collaborators.
- Create professional reports that document our workflow and results directly from our code, reducing the risk of accidental copy and paste or transcription errors.
- Document our workflow to facilitate reproducibility.
- Efficiently change code outputs (figures, files) given changes in the data, methods, etc.
The
knitrpackage was designed to be a transparent engine for dynamic report generation withR– Yihui Xi – knitr package creator
When To Knit: Knitting is a useful exercise throughout your scientific workflow. It allows you to see what your outputs look like and also to test that your code runs without errors. The time required to knit depends on the length and complexity of the script and the size of your data.
How to Knit
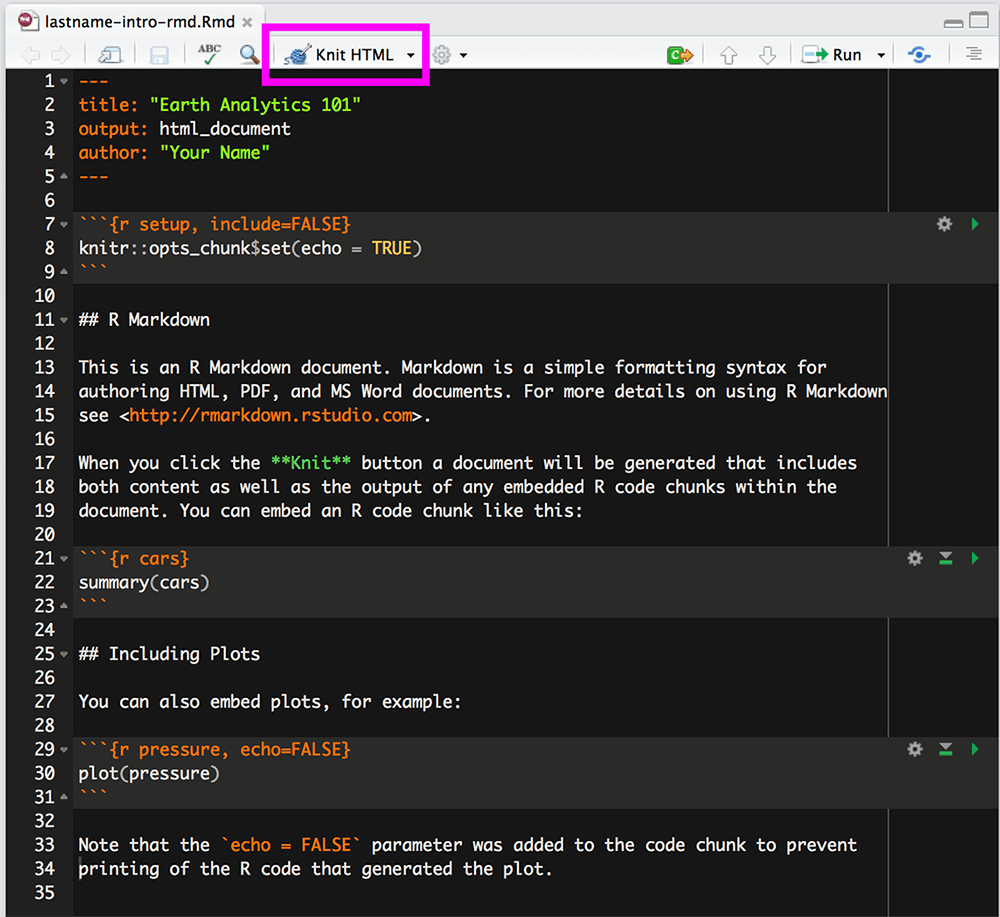
To knit in RStudio, click the Knit pull down button. You want to use the
Knit HTML option for this lesson.
When you click the Knit HTML button, a window will open in your console
titled R Markdown. This
pane shows the knitting progress. The output (html in this case) file will
automatically be saved in the current working directory. If there is an error
in the code, an error message will appear with a line number in the R Console
to help you diagnose the problem.
Data tip: You can run knitr from the command prompt
using: render(“input.Rmd”, “all”).
View the Output
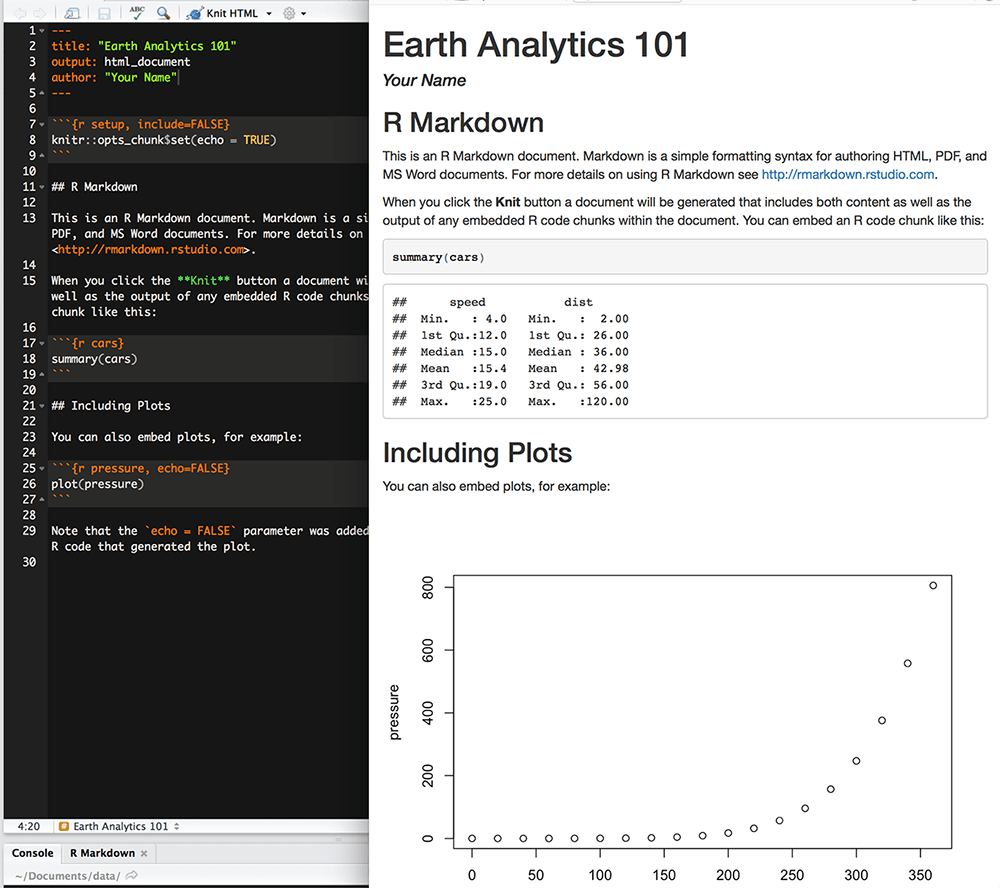
When knitting is complete, the html file produced will automatically open.
Notice that information from the YAML header (title, author, date) is printed
at the top of the HTML document. Then the html shows the text, code, and
results of the code that you included in the Rmd document.
Challenge Activity
Add the code below to your .Rmd document. Then knit to .html format.
# load the ggplot2 library for plotting
library(ggplot2)
# download data from figshare
# note that we are downloading the data into your working directory (earth-analytics)
download.file(url = "https://ndownloader.figshare.com/files/7010681",
destfile = "data/boulder-precip.csv")
# import data
boulder_precip <- read.csv(file="data/boulder-precip.csv")
# view first few rows of the data
head(boulder_precip)
# when we download the data we create a dataframe
# view each column of the data frame using it's name (or header)
boulder_precip$DATE
# view the precip column
boulder_precip$PRECIP
# q plot stands for quick plot. Let's use it to plot our data
qplot(x=boulder_precip$DATE,
y=boulder_precip$PRECIP)
When you knit your .Rmd file to pdf, the plot you produce should look like
the one below. Not so pretty, eh? Don’t worry - we will learn more about plotting
in a later tutorial!

Where is the File?
In the steps above, we downloaded a file. However, where did that file go on your computer? Let’s find it before we go any further.
# what is the working directory?
getwd()
[1] "/Users/lewa8222/Documents/earth-analytics"
# set working dir as a variable
my.dir <- getwd()
# what files are in that working directory?
list.files(my.dir, recursive= TRUE)
Is the boulder-precip.csv file there?
Share on
Twitter Facebook Google+ LinkedIn
Leave a Comment